An AI system developed by Google’s DeepMind has made a “once in a generation” breakthrough that could have a dramatic impact on the way we treat diseases: accurately predicting how proteins fold into shapes.
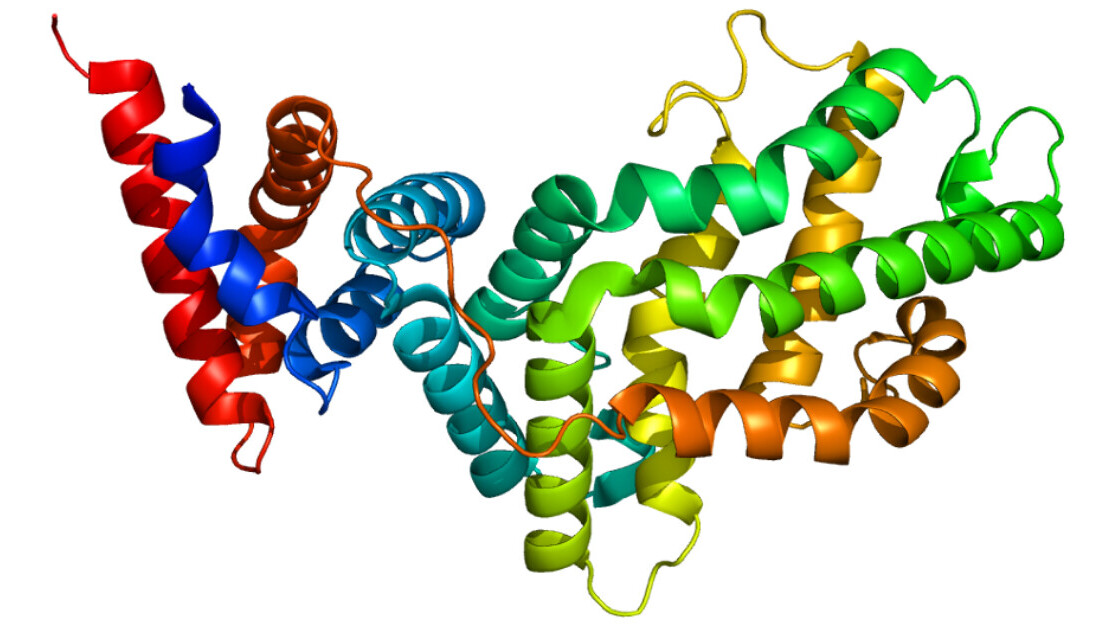
Proteins underpin almost every biological process. Each one is comprised of strings of amino acids that fold into countless elaborate shapes that determine what they do.
Predicting these structures can help us understand how proteins cause diseases — and develop drugs to treat them. But scientists currently know the exact 3D shapes of just a fraction of the 200 million-plus proteins they’ve identified.
[Read: How to build a search engine for criminal data]
Figuring out a protein’s precise form is typically a time-consuming and expensive process. But DeepMind says its AlphaFold system can predict their structures in a matter of days.
DeepMind tested its performance at the Critical Assessment of protein Structure Prediction (CASP) competition, a biannual experiment in predicting the 3D structure of proteins.
The London-based lab had won the 2018 edition of the challenge, but its predictions lacked the accuracy required to be biologically useful. This year, DeepMind revamped the system.
The latest version of AlphaFold uses a neural network system to identify the structure of a folded protein as a “spatial graph,” where residues are the nodes and edges connect the residues.
The system was trained on around 170,000 protein structures from the Protein Data Bank, and uses a relatively modest amount of computing power — roughly equivalent to 100-200 GPUs run over a few weeks.
In results from the latest CASP assessment released on Monday, two-thirds of the system’s predictions matched the accuracy of lab experiments, with an average margin of error comparable to the width of an atom.
“This computational work represents a stunning advance on the protein-folding problem, a 50-year-old grand challenge in biology,” said Professor Venki Ramakrishnan, the winner of the 2009 Nobel Prize for Chemistry. “It has occurred decades before many people in the field would have predicted. It will be exciting to see the many ways in which it will fundamentally change biological research.”
DeepMind is already exploring the possibilities. The company says it’s investigating how protein structure predictions could deepen our understanding of specific diseases, and believes they could be useful in future pandemic responses. In time, they could help find enzymes that break down plastic waste and even capture carbon from the atmosphere.
There’s still a long way to go before the scientific breakthrough is translated into real-world impacts. DeepMind admits that not all its predictions will be perfect, and still has much to learn about applying protein structures to practical purposes. But the work shows AI has the potential to deliver on some of the hype.
Get the TNW newsletter
Get the most important tech news in your inbox each week.